
DNA Fingerprinting of Date Palm Varieties (Phoenix dactylifera L.) Grown in Sudan Using ISSR Markers and SDS-PAGE
DNA Fingerprinting of Date Palm Varieties (Phoenix dactylifera L.) Grown in Sudan Using ISSR Markers and SDS-PAGE
Manal A Ibrahim1, Gamal E Osman2
1Department of Botany, Faculty of Science and Technology, Omdurman Islamic University, Sudan.
2Biology Department, Genetic Molecular Laboratory, Imam Abdalruhman bin Faisal University, Saudi Arabia
| ARTICLE INFORMATION | ABSTRACT | |
| Corresponding author: E-mail: manalabdalla071@gmail.com; manalabdalla@oiu.edu.sd Keywords: Date palm Fingerprint ISSR SDS-PAGE UPGMA Received: 27.07.2023 Received in revised form: 03.08.2023 Accepted: 5.08.2023 | Date palm (Phoenix dactylifera L.) family (Arecaceae) is the most important and ancient cultivated species in Sudan. Protein-based (SDS-PAGE) and Inter- Simple Sequence Repeat (ISSR) PCR) were used to identify genetic distance and design the phylogenetic tree for the five different date palm varieties (Mishriq, Barhi, Khadrawi, Sagay, and Khenaizy). EightI SSR- PCR primers which were used to amplify DNA segments from five date palm varieties were annealed with 60 loci across all variety genomes with an average of 7 loci per primer with a range of 200 to 950bp. Among those loci scored, 51 loci were polymorphic with (85%) polymorphism for at least one of the varieties with an average of 6 polymorphic bands per primer. A total of 159 bands from all analyses with an average of 19.8 fragments per primer, were enough for the identification and evaluation of these five date palm varieties. According to ISSR analysis, UPGMA (Unweight Pair Group of Arithmetic Averages) classified the fifty-one polymorphic loci into two main clusters, the first one contained two varieties: Mishriq and Barhi. While Khenaizi, Sagay, and Khadhrawi grouped in the second one which consisted of two sub-clusters, the first one consisted of Khenaizi and Khadhrawi, and the second sub-cluster consisted of Sagay variety. The combined tree of ISSR and SDS-PAGE analysis classified the date palm varieties under study into two main clusters. The first one consisted of two varieties: Mishriq and Barhi, which were closely related varieties. While the second one consisted of two sub-clusters, the first one consisted of two varieties, Khenaizi and Sugay and the other sub-cluster contained of Khadhrawi variety. |
INTRODUCTION
Date palm (Phoenix dactylifera L.) is an angiosperm that belongs to monocots and is considered the most important and ancient cultivated species in the Arab world (Elshibli, 2009). Date Palm has a major socio- economic importance because of its high nutritional value, great yields and its long life span. Date palms in Sudan have been traditionally grown using old, local varieties, mainly of the dry type, for 3000 years. (Elshibli and Korpelainen, 2009).
Numerous males were used as the source of pollen for hand pollination of the female trees. (Osman and Boulos, 1978). Female trees are cultivated mainly for their nutritive fruits. Although the average economic life of a date palm tree is estimated to be up to 50 years, the tree may stay productive for up to 150 years (Chao and Krueger, 2007).
Date contains many nutrients such as carbohydrates, proteins, fat, minerals and vitamins (Al-Qarawi et al. 2004). It is a good source of high nutritional value food. Indeed it is rich in carbohydrates, dietary fibers, proteins, minerals, and vitamin B complex, such as thiamine (B1), riboflavin (B2), niacin (B3), pantothenic (B5), pyridoxine (B6), and folate (B9) (Eoin, 2016).
Carbohydrates form 70% of date fruit and are mostly fructose and glucose in equal ratio while date proteins are rich in amino acids that contain acidic side chains but poor in methionine and cysteine, which their side chain are composed of sulfur. Minerals in date fruits are calcium, iron, magnesium, selenium, copper, phosphorus, potassium, zinc, sulfur, cobalt, fluorine, manganese, and boron (Chao and Krueger, 2007; Al- Harrasi et al. 2014). Date fruits are highly nourishing and may have numerous potential health benefits. The protective effects of fruits against chronic diseases are ascribed to bioactive non-nutrients called phytochemicals. Phytochemicals have gained increased interest among several investigators, including clinicians due to their antioxidant activity, cholesterol-lowering properties, and other potential health benefits such as chemoprevention of cancer, prevention of diabetes and cardiovascular diseases (Chao and Krueger, 2007; Al-Harrasi et al. 2014).
DNA markers have proved valuable in crop breeding, especially in studies of genetic diversity and gene mapping, phylogenetic studies, gene tagging, genome mapping, and evolutionary biology in a wide range of crop species (Gupta and Varshney, 2000).
ISSRs markers are powerful tools to study the inter- and intra-specific genetic variations in date palms and an easy approach with highly reproducible and multiple genomic loci target ability. To assess the genetic diversity on the basis of geography, the genetic relationship among genotypes was inferred using UPGMA cluster analysis representing the closeness and divergence among date palm cultivars. The present study will be helpful for germplasm management in order to improve the conservation and production of elite cultivars.
MATERIALS AND METHODS
Plant materials
The green and yellow leaflets female samples were collected from EL Slate farm in north Khartoum (Barhi, Khenaizi, Khadhrawi, Sugay andMishriq Wad Laggai). Sagay has been recently introduced from the Kingdom of Saudi Arabia, (Khonaizi) from the United Arab
Emirates, (Khadhrawi and Barhi) from Iraq and Mishriq Wad Laggai which was domesticated in Sudan, the plant materials were collected and transferred in Liquid nitrogen (-190°C).
DNA extraction: (CTAB Method)
Cetyl trimethyl ammonium bromide (CTAB) method (Allen et al. 2006).) 50-mg samples of young leaflets tissues were ground to a fine powder in liquid nitrogen, the powder was then placed in .5- mL microtubes containing 700 µL 3% CTAB extraction buffer. The solution was incubated at 65ºC for 60 min, gently mixing by inversion every 15 min; an equal volume of chloroform-isoamylalcohol (24:1) was added to the tubes and gently mixed for 1 min. Samples were centrifuged for 10 min. at 10,000 rpm; 600 µL of the supernatant was then transferred to a fresh tube following the addition of 500 µL chloroform-isoamylalcohol (24:1); this procedure was repeated twice; 500 µL of the supernatant was then transferred to a fresh tube with 700 µL of cold isopropanol (-20ºC) and 1/10th volume of 3 M sodium acetate samples were gently mixed by inversion and centrifuged at 12,000 rpm for 10 min, and so it was possible to visualize the DNA adhered to the bottom of the tube. The liquid solution was then released and the DNA pellet was washed with 700 µL of 70% ethanol and set to dry for approximately 12 h, or until the next day, with the tubes inverted over a filter paper, at room temperature; the pellet was then re- suspended in 100 µL TE buffer plus 5 mL ribonuclease (RNase 10 mg mL–1) in each tube; this solution was incubated at 37ºC for 1h, and after stored at -20ºC. They were used as template DNA for ISSR primer analysis.
Qualitative and Quantitative Analyses of Extracted DNA
DNA yield was measured using a UV-visible spectrophotometer (PerkinElmer, Waltham, MA, USA) at 260nm. DNA purity was determined by calculating the absorbance ratio at A260/280nm. (Wilson and Walker, 2005). For quality and yield assessments, electrophoresis was performed for all DNA samples on 0.8% garose gels that were stained with ethidium bromide; the bands were observed and compared with a known standard lambda DNA marker sample.
ISSR-PCR
Primers Selection: three types of primers, produced by (Sigma Aldrich, Banglore),six anchored dinucleotides repeat primer (AG)10C, (AG)10T, (CT)10A, (CT)10T,
(CT)10G, and (CA)8GT, one nonanchored tetra- nucleotides repeat primer (GACA)4, and one nonanchored tri- nucleotides (CAG)5. Which were preselected for their performance with date palm DNA were tested to arrive at the primitive primer which gives descriptive segments (polymorphism, Table 1).
Table 1: The ISSR primers used in the study
| No | Primer | Sequence 5ʹ— 3ʹ | Annealing Temp. C0/Sec |
| 1 | DPISR1 | AGCAGCAGCAGCAG | 50.9 / 57.8 |
| 2 | DPISR2 | CACACACACACACACAGT | 50.9 / 57.8 |
| 3 | DPISR3 | GACAGACAGACAGACA | 48 / 47.5 |
| 4 | DPISR4 | AGAGAGAGAGAGAGAGAGAGC | 50.9 / 57.8 |
| 5 | DPISR5 | AGAGAGAGAGAGAGAGAGAGT | 50.9 / 57.8 |
| 6 | DPISR6 | CTCTCTCTCTCTCTCTCTCTA | 50.9 / 57.8 |
| 7 | DPISR7 | CTCTCTCTCTCTCTCTCTCTT | 50.9 / 57.8 |
| 8 | DPISR8 | CTCTCTCTCTCTCTCTCTCTG | 50.9 / 57.8 |
The polymerase chain reaction (PCR) Optimization
According to Williams 1990, the polymerase chain reaction (PCR) mixture (25 µl )consisted of 2 µl of total genomic DNA, 12.5 µl of Ampli Taq Gold 360 Mastermix (Applied Biosystems), 2 µl (5 pmol/ µl) of each primer and 8.5 µl of nuclease-free water. Amplification took place in DNA amplification The rmocycler (Biorad, icycler), is programmed as a denaturation step of 4 min at 94 ºC followed by 35 cycles compose of 30 seconds at 94 ºC, for 30 seconds at annealing temperature and 3 minutes at 72ºC. a final extension of 72 ºC for 5 minutes, and hold at 4ºC.
Agarose Gel Electrophoresis
Amplification products were electrophoresed on 1.8% agarose gel (Sigma) in 100 mil 0.5XTBE buffer. The gel was run at 120V constant voltages for 45 minutes. The 100 bp standard DNA size marker (ladder) (Sigma Aldrich, Banglore) was run along with the samples to compare the molecular weight of amplified products. Gels were stained with 0.5 μg/mL ethidium bromide for 15 min (Caetano-Anolles, 1997).
Visualization and analysis of PCR products
Visualization of amplification products and data analysis of reproducible bands visualized on agarose gels 1.8% were scored using a binary code in a data matrix 1 and 0 for their present and absent respectively for the eight primers. Fragments with the same mobility were considered identical, irrespective of the intensity of the fragment.
Statistical Analysis
ISSR, SDS-PAGE, and combined analysis data were converted into binary data in an Excel worksheet and were analyzed using the SPSS-16 program to find the genetic distance between and within the five different date palm varieties. Unweight Pair Group Method with Arithmetic Average (UPMGA) analysis was used for cluster analysis using ISSR, data based on the Jaccard similarity matrix which were computed with the SPSS-10 program to produce a genetic distance matrix using Dice similarity coefficients19. A dendrogram was generated by cluster analysis using the unweighted pair group method of the arithmetic averages (UPGMA).
RESULTS AND DISCUSSION
ISSR polymorphism
The results obtained through eight ISSR primers (as listed in Table 2 and Fig 1) showed the eight ISSR-PCR primers were used to amplify DNA segments from five date palm varieties (Khenaizi, Sugay, Khadhrawi, Mishriq and Barhi), were annealed with 60l ocia cross all variety genomes with an average of 7l ociperprimer with a range of 200 to 950 bp. Among those lociscored, 51l oci were polymorphic with (85%) polymorphism for atleast one of the varieties with an average of 6 polymorphic bands per primer.
A total of 159 bands from all analysis with an average of 19.8 fragments per primer, which were enough for the identification and evaluation of genetic diversity and designing the phylo genetic tree for these five different date palm varieties. These results are in agreement with those of Zehdi et al. (2004), using ISSR on Tunisi a date palm that generated 100 bands were identified at 14 microsatellite loci with average of
7.14 all elesperlocus. Adawy et al. (2002) using seven ISSR primers generated 53 fragments ranging from 298 to 1200 bp in size.
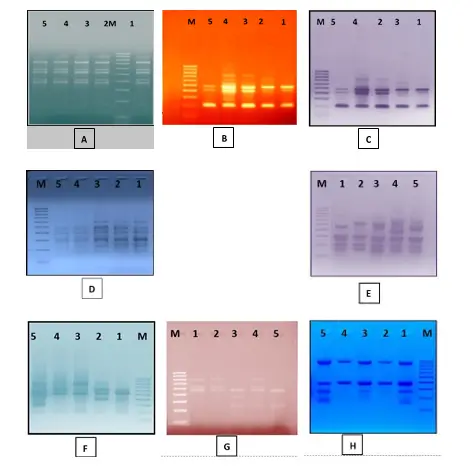
Fig.1: ISSR profile of five date palm varieties amplified with eight different ISSR primers. (A)Primer DPISR-1, (B) Primer DPISR-2, (C)DPISR-3 (D)DPISR-4 ,(E)DPISR-5(F)DPISR-6,(G)DPISR-7and,(H) DPISR-8.M: 100bp ladder
marker. Lanes 1 through 5 refer to date palm varieties Khenaizi, Sugay, Khadhrawi, Mishriq and Barhi, respectively.
The average number of fragments per primer was 7.6 fragments with 64.1%polymorphism. While, Adawy et al. (2004) generated 159 fragments when using 19 ISSR primers to analyze bulked DNA samples representing five date palm varieties and the average number of fragments/primer was 8.4.
In Table:4.6 the number of amplified fragments per primer varied from 13bands for the primer DPISR7, which showed the lowest primers efficiency(8.2 %,) to 29 bands for the primer DPISR-1 which represented the highest efficiency (18.2 %,) Table 3.19 The most informative primers, considering the percentage of polymorphism (% P = 100) were DPISR-5, DPISR-6 and DPISR-8 with 12, 8 and 7 polymorphic bands respectively indicating their abundance over other in date palm genome while the non –anchored (CAG)tri-(DPISR1) nucleotides exhibited the lowest polymorphism (16.6 %).
On the other hand polymorphic band with discrimination power (2%) indicated the rareness of such repeat among the five varieties analyzed. This is due to the absence of sites that complement the sequences of these primers in the palm genome and the extent of polymorphism varies with the nature of the primer used and the sequence of repeats (motif) in the primer employed. This result together with those obtained by Ruas (2003) and Perezdela Torre et al. (2010) indicates that the level of polymorphism detected by ISSR primers depends on the species or genus and the repetitive ISSR used in the primer utilized to generate the amplification profiles, additionally, this result is in agreement with what was reported by Zhao et al. 2012 who stated that the AG– ISSR repeat is the most abundant and polymorphic among di-nucleotide and comprises 85.7 % of date palm genome.
Table 2: List of the used primers and the complementary information of the ISSR assay
| Primer | No. ofloci | NMB * | NPB * | TBN * | P%* | %E * | %D * | ABL(bp) * |
| DPISR1 | 6 | 5 | 1 | 29 | 16.6 | 18.2 | 2.0 | 380-900 |
| DPISR2 | 6 | 1 | 5 | 16 | 83.3 | 10 | 9.8 | 200-700 |
| DPISR3 | 6 | 1 | 5 | 19 | 83.3 | 12 | 9.8 | 200-800 |
| DPISR4 | 10 | 1 | 9 | 24 | 90 | 15.1 | 17.6 | 200-600 |
| DPISR5 | 12 | 0 | 12 | 26 | 100 | 16.3 | 23.5 | 200-580 |
| DPISR6 | 8 | 0 | 8 | 16 | 100 | 10.1 | 15.7 | 280-850 |
| DPISR7 | 5 | 1 | 4 | 13 | 80 | 8.2 | 7.8 | 350-800 |
| DPISR8 | 7 | 0 | 7 | 16 | 100 | 10.1 | 13.7 | 200-950 |
| Total Average | 60 7 | 9 1.2 | 51 6 | 159 19.8 | 85 81.7 | 100 12.5 | 100 12.5 |
*TBN: Total band number, NPB: Number of polymorphism bands, NMB: Number of monomorphism bands, P%: Polymorphism percentage, ABL: The amplified band length, D%: Discrimination power, E%: Primer Efficiency.
It is clear that the ISSR markers differed among them in the number of bands according to the marker. This was shown by several studies, Karim el al. (2010), studied the genetic convergence between the ten date palm varieties in Tunisia to find the genetic convergence between the ten date palm varieties they used the same primers, and only seven of them gave a resultwith11bandsperprimer (AG10C, AG10T, CT10A, CT10T, CT10A, CT10G and GACA4).
Genetic distance and phylogenetic tree analysis
In the present study ISSR, SDS-PAGE and combined analysis data were converted into binary data and were analyzed using the SPSS-16 program to construct the genetic distance between the five different date palm genotypes (Table 3, 4 and 5). Three phylogenetic trees that were generated showed six similar clusters (Fig. 2, 3 and 4).
The genetic distances matrix was calculated for the 51 polymorphic bands of ISSR, 8 protein patterns and combined analysis of the five varieties on the basis of present and absent of the polymorphic bands. The genetic distance and separation of each variety varied according to the type of analysis used. The range is between (0.922-0.645 Table 3), (0.816- 0.289 Table 4)
and (0.887-0.627 Table 5) with a mean of (0.783- 0.552-0.757) for the three analyses respectively. Thus the genotypes that tested in this study are highly divergent at the DNA level.
The smallest distance value observed between Khenaizi and Sagay varieties for the SDS-PAGE and combined-based analysis was 0.289, and 0.627 respectively, which appear to be the most similar varieties and can be closely related. The Sugay variety was highly divergent from Barhi variety with a distance of 0.922 – 0.887 for the ISSR and combined-based analysis respectively.
It is noteworthy that Mishriq presented a very limited average distance (from 0.780 to 0.849) with Khenaizi, Sugay and Khadhrawi, thus Mishriq could be characterized by a high divergence at the DNA level and could be unlikely to regroup with them. All the varieties displayed different intermediate levels of dissimilarity (0.627 to 0.658) and are grouped with the other ones.
Table 3: Genetic distance values of Nei’s coefficient revealed by ISSR markers analysis
| Matrix File Input | |||||
| Khenaizi | Sagay | Khadhrawi | Mishriq | Barhi | |
| Khenaizi | .000 | ||||
| Sagay | .664 | .000 | |||
| Khadhrawi | .645 | .683 | .000 | ||
| Mishriq | .852 | .774 | .751 | .000 | |
| Barhi | .797 | .922 | .659 | .761 | .000 |
Table 4: Genetic distance values of Nei’s coefficient revealed by SDS- PAGE analysis
| Matrix File Input | |||||
| Khenaizi | Sagay | Khadhrawi | Mishriq | Barhi | |
| Khenaizi | .000 | ||||
| Sagay | .289 | .000 | |||
| Khadhrawi | .516 | .408 | .000 | ||
| Mishriq | .548 | .577 | .816 | .000 | |
| Barhi | .447 | .500 | .707 | .408 | .000 |
Table 5: Genetic distance values of Nei’s coefficient using combined data of ISSR and SDS-PAGE analysis
| Matrix File Input | |||||
| Khenaizi | Sagay | Khadhrawi | Mishriq | Barhi | |
| Khenaizi | .000 | ||||
| Sagay | .627 | .000 | |||
| Khadhrawi | .637 | .658 | .000 | ||
| Mishriq | .849 | .780 | .793 | .000 | |
| Barhi | .780 | .887 | .701 | .749 | .000 |
The trees constructed were shown in Fig 2, 3 and 4 explained the molecular phylogenetic relationships between the five varieties. Unweight Pair Group Method with Arithmetic Average (UPMGA) analysis classified date palm varieties into two main clusters in ISSR, SDS-PAGE, and ISSR, SDS-PAGE combined analyses.
Fifty-one polymorphic loci, UPGMA classified the date palm varieties into two main clusters according to combined analyses that were revealed by ISSR and SDS-PAGE combined analyses. The first one consisted of two varieties: Mishriq and Barhi it is similar to those based on agronomic traits (Fig 4). And the second one consisted of two sub-clusters, the first one consisted of two varieties, Khenaizi and Sagay and the second sub-cluster consisted of Khadhrawi variety (Fig. 4).
UPGMA ordered the 51 polymorphic loci of ISSR analysis of the date palm varieties into two main clusters the first one contained two varieties: Mishriq and Barhi. While Khenaizi, Sugay and Khadhrawi grouped in the second one which consisted of two sub-clusters, the first one consisted of Khenaizi and Khadhrawi, and the second sub-cluster consisted of Sagay variety (Fig 2).
The combined tree of ISSR and SDS-PAGE analysis in (Fig.4) classified the date palm varieties under study into two main clusters according to UPGMA analysis similar to the SDS-PAGE analysis tree (Fig 3). The first one consisted of two varieties: Mishriq and Barhi, which were closely related varieties. While the second one consisted of two sub-clusters, the first one consisted of two varieties, Khenaizi and Sagay and the second sub-cluster consisted of Khadhrawi variety.
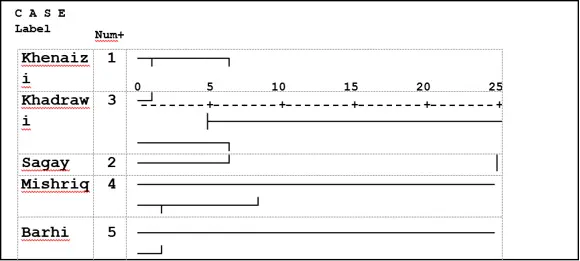
Fig 2: Cluster analysis with UPGMA method of five date palm varieties using ISSR, data based on Jaccard similarity matrix.
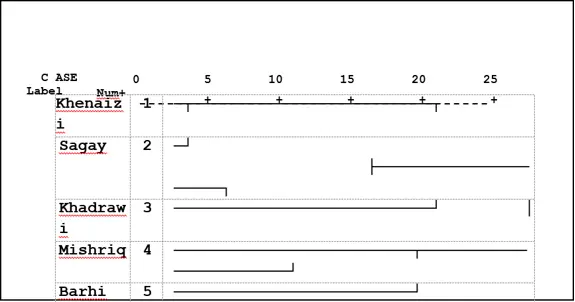
Fig 3: Cluster analysis with UPGMA method of five date palm varieties using SDS-PAGE data based on Jaccard similarity matrix.
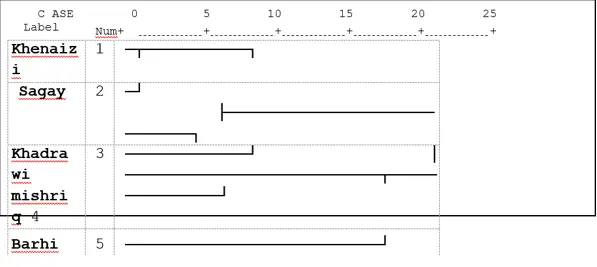
Fig 4: Cluster analysis with UPGMA method of five date palm varieties using combined data of ISSRand SDS- PAGE based on Jaccard similarity matrix.
Generally, the genotypes tested revealed that the geographic origin was not affected the cluster divisions. Accordingly, the sister varieties `Khenaizi` and ‘Sagay with (0.289, 0.627 distances Table 2 and 3) which have different geographical origins (The United Arab Emirates and the Kingdom of Saudi Arabia respectively) fell in one sub-cluster (Fig 4 and 3). Also, the two varieties Mishriq and „Barhi‟ (Sudan and Iraq, respectively) fell in the same cluster, This result agrees with other reports for Moroccan, Algerian and Tunisian date palm varieties based on analyses using microsatellite markers (Zehdi et al. 2004) and isozyme markers (Ould et al. 2001).
A high degree of independence between the geographical origin and molecular data was indicated. The RAMPO and AFLP data applied on Tunisian date palms (Rhouma et al. 2007; Rhouma-Chatti et al. 2011) showed that the studied varieties clustered independently of their geographic origin. The two varieties Mishriq and Barhi (Sudan and Iraq, respectively) which grouped into the same cluster, based on agronomic traits particularly the fruits which were characterized by dates of medium size and brown color. This could be explained by the presence of a common genetic origin among the tested varieties in spite of their origin and this agrees with Hammadi et al. (2012) who found that fruit consistency which is an important characteristic of date fruit has an association with genetic markers because clustering based on fruit consistency is in accordance with clustering by microsatellite markers.
The varieties Barhi and ‘Khadhrawi originated in Iraq and were grouped in a different cluster, this observation suggested that genetic variation range with each of the Iraqi varieties exits grouped, this result confirmed what was obtained by Al-Najm et al. (2017) when they used inter-primer binding site (iPBS) markers to assess the molecular variation and genetic diversity of 54 and 12 date palm genotype collected from Australia and Iraq, they found that Barhi and ‘Khadhrawi originated in Iraq in a different group to those collected in Iraq , their observations suggests that a range of genetic variation within each of the Iraqi varieties exists. So this could be helpful in Sudan where date palm breeding is highly dependent on seed propagation with subsequent selection based on specific characteristics such as fruit quality and plant vigor as determined by local farmer preferences (Khierallah et al. 2011). Results also showed that the imported date palm varieties recently introduced to Sudan groves are closely grouped in one cluster this could be explained by the presence of a common genetic origin among them, Al-Khalifah et al., (2013), added that over the years many date palm varieties have been transplanted to areas other than the area of their origin, and there may have been adapted with different names. On the whole, our data augment those describing the application of molecular tools in date palm variability analysis and previously reported (Trifi et al. 2000). Dendrogram showed that accession grouping in relation to their geographical origin is not well defined.
ISSR data allowed the discrimination of five varieties. However, the use of SDS- PAGE, in spite of the low numbers of bands could distinguish Khadhrawi and Mishriq varieties by negative unique bands, while ISSR assay could distinguish the five varieties through 16 unique bands.
CONCLUSION
The combined cluster analysis of ISSR and SDS-PAGE data clearly showed a high degree of independence between the geographical origin and molecular data which were indicated. The imported date palm varieties recently introduced to Sudan are closely grouped in one cluster this could be explained by the presence of a common genetic origin among them. Generally, the genotypes tested revealed that the geographic origin did not affect the cluster divisions. Moreover, ISSR profiles can be used in developing molecular identities for date palm varieties in order for their proper identification, registration, and conservation.
ACKNOWLEDGMENT
The authors are highly thankful and deeply grateful to members of the Biology Department, Molecular Laboratory, Imam Abdulrahman bin Faisal University, Saudi Arabia, for their support and materials during the study.
REFERENCES
Abdulla, M.; Gamal O. Investigation on molecular phylogeny of some date palm (Phoenix dactylifera L.) varieties by protein, RAPD and ISSR markers in Saudi Arabia. Aust. J Crop Sci., 2010, 4: 23–28.
Adawy, S.S.; Hussein Ebtissam, H.A.; ElKhishin D.; Moharam, H.; ElItriby, H.A. Genetic variability studies and molecular fingerprinting of some Egyptian date palm (Phoenix dactylifera L.) varieties: II. RAPD and ISSR profiling. Arab J. Biotech., 2002, 5(2), 225-236.
Adawy, S.S.; Hussein, E.H.A.; Saker, M.M.; El-Itriby, H.Intra- and Inter-varietal variation of Upper Egypt date palm varieties (Phoenix dactylifera L.): I. As revealed By RAPD and ISSR markers. Proceed. Int. Conf. Genet. Eng. & Appl., Sharm El-Sheikh, South Sinai, Egypt, 2004, (8)11, 165-
179.
Al-Harrasi, A.; Rehman, N.U.; Hussain, J.; Khan, A.L.; Al-Rawahi, A.; Gilani, S.A. et al. Nutritional assessment and antioxidant analysis of 22 datepalm (Phoenix dactylifera) varieties growing in Sultanate of Oman. Asian Pac. J.Trop. Med., 2014, (7) 2, 591–598.
Al-Khalifah, N.S.; Askari, E.; Shanavaskhan, A. Date palm tissue culture and genetically identification of varieties grown in Saudi Arabia. National Center for Agriculture Technologies, King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia., 2013.,207.
Al-NajmA, L.S,; Ahmad, N.M.; Pourkheirandish, M.; Trethowan, R. Molecular svariability and population structure of a core collection of date palm (Phoenix dactylifera L.) varieties from Australia and the Middle East. AJCS, 2017, 11(09),1106-1115.
Al-Qarawi, A.A.; Mousa, H.M.; Ali, B.H.; Abdel- Rahman, H.; El- Mougy, S.A. Protective effect of extracts from dates (Phoenix dactylifera L.) on carbon tetrachloride–induced hepatotoxicity in rats. Inter. J. Appl. Res. Vet. Med., 2004, 2, 176–180.
Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc, 2006, 1:2320– 2325.
Arslan, E,; Tamkoc, A. The application of ISSR-PCR to determine the genetic relationship and genetic diversity between narrow leaved bluegrass (Poa angustifolia) and rough bluegrass (Poatrivialis) accessions. Turk J Biol., 2011, 35, 415–423.
Caetano-Anolles, G. Resolving DNA amplification products using polyacrylamide gel electrophoresis and silver staining. In: Fingerprinting Methods based on Arbitrarily Primed PCR, 1997, 119‒134. Springer, Berlin, Heidelberg, Germany
Chao, C.T.; Krueger, R.R. The date palm (Phoenix dactylifera L.): overview of biology, uses, and cultivation. Hort. Science, 2007, 42(5), 1077-1082.
Elshibli, S,; Korpelainen, H. “Biodiversity of date palms (Phoenix dactylifera L.) In Sudan: chemical, morphological and DNA polymorphisms of selected varieties,” Plant Genetic Resources: Characterization and Utilization, 2009, (7), 2,194–203.
Elshibli.S.; Korpelainen, H. “Biodiversity of date palms (Phoenix dactylifera L.) In Sudan: chemical, morphological and DNA polymorphisms of selected varieties,” Plant Genetic Resources:
Characterization and Utilization., 2009, 7(2), 194–203.
Eoin, L.N. Systematics: blind dating. Journal of Nat.
Plants, 2016, 2, 69.
Gupta, P,K.; Varshney, R.K. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica, 2000, 113,163–185.
Hammadi, H.; Benabderrahim, M.A.; Elbekkay, M.; Ferdaous, G.; Triki, T.; Ferchichi, A. Investigation of genetic variation in Tunisian date palm (Phoenix dactylifera L.) varieties using ISSR marker systems and their relation with fruit characteristics. Turk J Biol., 2012, 36, 449-458.
Karim, K.; Chokri, B.; Amel. H.; Wafa, H.; Richid, H.; Nouredine, D. Genetic diversity of Tunisian date palm germplasm using ISSDR markers. Int. J. Bot., 2010, 6(2), 182-186.
Khierallah, H.; Bader, S.; Baum, M.; Hamwieh, A. Assessment of genetic diversity for some Iraqi date palms (Phoenix dactylifera L.) using amplified fragment length polymorphisms (AFLP) markers. Afr. J. Biotechnol., 2011, 10, 9570-9576.
Osman, A.M.A.; Boulos, N.N. Evaluation of some Sudanese date varieties. Acta. Hortic., 1978, 185-190.
Ould Mohamed, S.A.; Trifi, M.; Sakka, H.; Rhouma, A.; Marrakchi, M. Genetic inheritance analysis of four enzymatic systems in date palm (Phoenix dactylifera L.). Genet Resour Crop Ev., 2001, 48, 361–368.
Perezdela, T.M.; Zirilli, P.; Ulrich, N.; Setten, L.; Escandon, A. Caracterización molecular en el género Mecardonia Ruiz & Pav.(Plantaginaceae) utilizando marcadores ISSR. Revista de la Facultad de Agronomía, La Plata, 2010, 109(1), 23-30.
RhoumaS, Z.S.A.; Ould, M.S.A.; Rhouma, A.; Marrakchi, M.; Trifi, M. Genetic diversity in ecotypes of Tunisian date palm (Phoenix
dactylifera L.) assessed by AFLP markers. J Hortic. Sci. Biotech., 2007, 82, 929–933.
Rhouma-Chatti, S.; Baraket, G.; Daskhlaoui-Dkhil, S.; ZehdiAzouzi, S.; Trifi, M. Molecular research on the genetic diversity of Tunisian date palm (Phoenix dactylifera L.) using the random amplified microsatellite polymorphism (RAMPO) and amplified fragment length polymorphism (AFLP) methods. Afr. J. Biotechnol, 2011, 10, 10352–10365.
Ruas, P.M.; Ruas, C.F.; Rampim, L.; Carvalho, V.P.; Ruas, E.A.; Sera,T. Genetic relationship in Coffea species and parentage determination of interspecific hybrids using ISSR (Inter- Simple Sequence Repeat) markers. Genetics and Molecular Biology, 2003, 26(3), 319-327.
TrifiM, R.A.; Marrakchi, M. Phylogenetic relationships in Tunisian date palm (Phoenix dactylifera L.) germplasm collection using DNA amplification fingerprinting. Agronomie, 2000, 20, 665–671. Williams, J.F. Optimization strategies for the polymerase chain reaction. Bio-techniques, 1990, 7, 762–9.
Wilson, K.; Walker, J. Principles and Techniques of Biochemistry and Molecular Biology. Cambridge University Press, Cambridge., 2005. Wilson, K.; Walker, J. Principles and Techniques of Biochemistry and Molecular Biology., Cambridge University Press, Cambridge, 2005.
Zehdi, S.; Trifi, M.; Billotte, N. et al. Genetic diversity of Tunisian date palms (Phoenix dactylifera L.) revealed by nuclear microsatellite polymorphism. Heredity, 2004, 141, 278–287.
ZehdiS, T.M.; Ould, M.S.A.; Rhouma, A.; Marrakchi, M. Survey of inter simple sequence repeat polymorphisms in Tunisian date palms (Phoenix dactylifera L.). J Genet Breed, 2002, 56, 77–83.
Zhao, Y.; Williams, R.; Prakash, C.S.; He, G. Identification and characterization of gene- based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol, 2012, 12, 237.

